Andhra Pradesh BIEAP AP Inter 2nd Year Botany Study Material 11th Lesson Biotechnology: Principles and Processes Textbook Questions and Answers.
AP Inter 2nd Year Botany Study Material 11th Lesson Biotechnology: Principles and Processes
Very Short Answer Questions
Question 1.
Define Biotechnology.
Answer:
It is a science which utilizes properties and uses of micro-organisms or exploits cells and the cell constituents at the industrial level for generating useful products essential to life and human welfare.
Question 2.
What are molecular scissors? Where are they obtained from?
Answer:
Molecular scissors are the restriction enzymes which cut the DNA at specific locations. Usually, they are obtained from Bacteria.
Question 3.
Name any two artificially restructured plasmids.
Answer:
PBR322, PUC19 101.
Question 4.
What is E coRI? How does it function?
Answer:
E.coRI is a restriction enzyme obtained from Escherichia coli. It specifically recognises GAA sites on the DNA and cuts it between G and A.
![]()
Question 5.
What are cloning vectors? Give an example.
Answer:
Vectors used for multiplying the foreign DNA sequences are called cloning vectors.
Ex: Plasmids, Bacteriophages, Cosmids.
Question 6.
What is recombinant DNA?
Answer:
The hybrid DNA formed by the fusion of DNA with desirable genes with the vector DNA by DNA ligase is called r-DNA.
Question 7.
What is palindromic sequence?
Answer:
The specific nucleotide sequence of DNA which is recognised by restriction enzyme is called palindrome, which is 4-6 base pairs in length.
Question 8.
What is the full form of PCR? How it is useful in Biotechnology?
Answer:
PCR stands for Polymerase chain reaction. It can be used for the diagnosis of diseases like AIDS, middle ear infection, and tuberculosis.
Question 9.
What is down stream processing?
Answer:
Separation and purification of products before they are ready for marketing is called
downstream processing.
Question 10.
How does one visualize DNA on an agar – gel?
Answer:
To make the DNA visible in the gel, Ethidium bromide is added to the gel solution and the buffer. This positively charged polycyclic compound binds to .DNA inserting itself between the base pairs. Southern blotting may also be used as visualization technique for agarose gels.
![]()
Question 11.
How can you differentiate between exonucleases and endonucleases?
Answer:
| Exonuclease | Endonuclease |
| 1. They remove nucleotides from the ends of the DNA | 1. They make cuts at specific locations with in the DNA |
Short Answer Questions
Question 1.
Write short notes on restriction enzymes.
Answer:
Two enzymes responsible for restricting the growth of Bacteriophage in Escherichia coli were isolated in the year 1963. One of these added methyl groups to DNA and the other cut DNA. The latter was called restriction endonuclease. The first restriction endonuclease – Hind II which cut DNA molecules at a particular point by recognising a specific Sequence of six base pairs, called recognition sequence for Hind II. Today, more than 900 restriction enzymes were isolated from over 200 strains of Bacteria, each of which recognises a different recognition sequence.
E CORI is a restriction enzyme in which, the first letter comes from the Genus (Escherichia), and the second two letters from the species of the Prokaryotic cell [coli], the letter ‘R’ is derived from the name of strain. Roman numbers indicate the order in which the enzymes were isolated from that strain of Bacteria. Restriction enzymes belong to a larger class of enzymes called nucleases. They are of two types.
a) Exonucleases which remove nucleotides from the ends of the DNA.
b) Endonucleases which make cut’s at specific location with in the DNA-
Question 2.
Give an account of amplification of gene of interest using PCR.
Answer:
PCR stands for Polymerase chain reaction. In this reaction, multiple copies of the gene of interest are synthesized in vitro using two sets of primers(oligonucleotides) and the enzyme DNA polymerase. The enzyme extends the primers using the nucleotides provided in the reaction and the. genomic DNA as template. As the process of replication of DNA is repeated many times, the segments of DNA can be amplified to approximately billion times.
Such repeated amplification is achieved by the use of a thermostable DNA polymerase such as Taq polymerase (isolated from thermus aquaticus) which remain active even during high temperature induced denaturation of ds DNA. The amplified fragment, if desired can: now be used to ligate with a vector for further cloning.
Question 3.
What is a bio-reactor? Describe briefly the stirring type of bio-reactor.
Answer:
Bio-reactor is a large vessel which is used for biological conversion of raw material into specific product.
A stirred-tank bio-reactor is usually cylindrical or with a curved base to facilitate the mixing of the reactor contents.
The stirrer facilitates even mixing and oxygen availability throughout the bio-reactor. Alternatively air can be bubbled through the reactor. It has an agitator system, an oxygen delivery system, a foam control system, a temperature .control system, pH control system and sampling ports, so that small volumes of the culture can be with drawn periodically.
![]()
Question 4.
What are the different methods of insertion of recombinant DNA into the host cell?
Answer:
There are several methods of introducing the ligated DNA into recipient cells. Recipient cells after making them competent to receive, take up the DNA present in their surrounding. R-DNA can be forced into such cells by incubating the cells with r-DNA on ice followed by placing them briefly at 42eC -(heat shock) and then putting them back on ice. This enables the bacteria to fake up the r-DNA.
In Micro-injection method, r-DNA is directly injected into the nucleus of an animal cell.
In Bio listic of gene gun method – cells are bombarded with high velocity micro particles of gold or tungsten coated with DNA.
In another method, ‘Disarmed pathogen’ vectors are used which when allowed to infect the cell, transfer the r-DNA into the host.
Long Answer Questions
Question 1.
Explain briefly the various processes of recombinant DNA technology.
Answer:
The important methods in recombinant DNA technology are performed through genetic engineering. They are :
i) Isolation of a desired gene,
ii) Insertion of isolated gene into a suitable vector,
iii) Introduction of recombinant vector into a host and
iv) Selection of the transformed host cells.
I) Isolation of a desired gene :
- The desired gene is isolated from the donor cell. Normally bacteria are the source of desired genes.
- The cell walls of bacteria are degraded with the help of enzymes.
- The cell membranes are lysed with the help of detergents.
- By treating the cellular constituents with phenols and suitable nucleases and by subjecting to gradient centrifugation, pure DNA is isolated.
- The purified DNA is cut into a number of fragments by restriction endonucleases.
- The restriction enzymes cleave DNA molecules in two ways.
i) In one way they cut both strands of DNA at exactly opposite points to each other. This results in DNA fragments with blunt ends or flush ends, where two strands end at the same point. Such cut is generally termed as even cut.
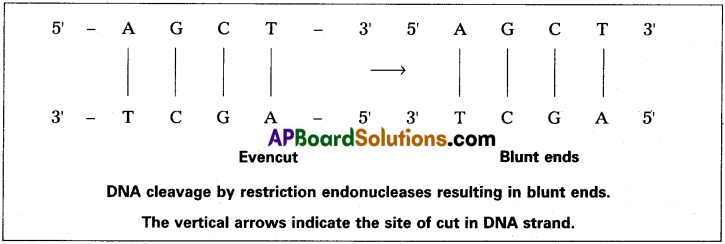
ii) But commonly, most enzymes cut the two strands of DNA double helix at different locations. Such a cleavage is generally termed as staggered cut. This generates protruding ends i.e., one strand of DNA double helix extends some bases beyond the other. Since the target site is palindromic in nature, the protruding ends generated by such a cleavage have complimentary base sequence.
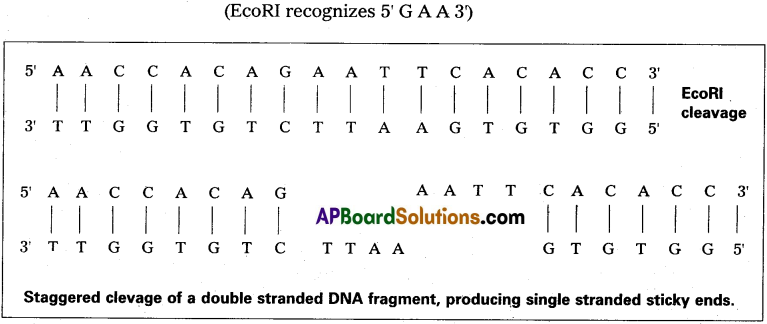
As a result, they readily pair with each other and such ends are called cohesive or sticky ends. This stickyness of the ends facilitates the action of the enzyme DNA ligase. When cut by the same restriction enzyme, the resultant DNA fragments have the same kind of ’sticky ends’ and these can be joined together readily by using DNA ligases. E.g.: The restriction enzyme E coRI.
E – The first letter, represents the name of genus Escherichia.
Co – The next two letters, represent the species Escherichia Coli.
The letter R is derived from the name of strain.
Roman numbers following the names indicate the order in which the enzymes were isolated from the strain of bacteria.
This enzyme specifically recognises GAA sites on the DNA and cuts it between G and A (G ↓ A)
7) The resultant fragments are separated from each other by gel electrophoresis.
8) The desired fragments are selected by Southern blotting technique.
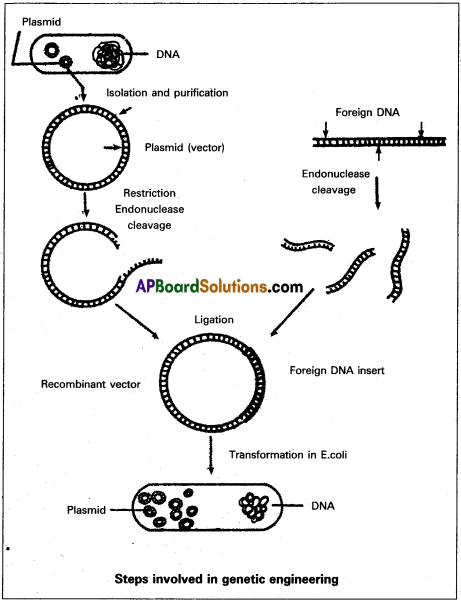
II) Insertion of isolated gene into a suitable vector :
- The selected fragments of DNA are inserted into a suitable vector to produce a large number of copies of genes. This is called gene cloning.
- There are two major types of vectors, namely plasmids and bacteriophages.
- Among the two types, plasmids are the ideal cloning vectors.
- To isolate a plasmid, the Bacterial cell is treated with EDTA (Ethylene diamine tetra acetic acid) along with lysozyme enzyme to digest the cell wall.
- Then the bacterial cell is subjected to centrifugation in sodium lauryl sulphate to separate the plasmid.
- The plasmid DNA is cut with the help of restriction endonuclease.
- The circular plasmid is converted into a linear molecule having sticky ends.
- The two sticky ends of linear plasmid are joined to the ends of desired gene by DNA ligase.
- The plasmid containing foreign DNA segments is called recombinant DNA (r DNA) or Chimeric DNA.
III) Introduction of recombinant vector into a suitable host :
- The r DNA molecule is introduced into suitable bacterial host cell by transfor-mation.
- The cell containing r DNA is called transformed cell.
- Bacterial cell walls are not permeable to recombinant vectors, but keeping in dil. Calcium chloride renders the bacterial cell wall permeable to recombinant vectors.
- The r DNA replicates with in the host cell.
- The transformed cell grows on the culture medium. Each daughter cell contains r DNA.
IV) Selection of transformed host cells :
1) Selection of transformed cells depends on the nature of gene which is cloned. 2) It can be done in two ways. The are :
a) Without using probes,
b) By using probes.
a) Without using probes:
- If the gene is cloned for antibiotic resistance, the cells are first incubated on a medium without antibiotic for one hour, to allow the antibiotic resistance gene to be expressed.
- Then the cells are placed on a medium with an antibiotic for selection of colonies containing rDNA.
- The cells which have expressed the gene will survive and the others die.
b) By using probes:
When transformed cells are cultured on the nutrient medium, several cells are produced. To select the cells containing the desired gene colony hybridization method is used. In this gene specific probes are used. A probe is a small fragment of single stranded RNA or DNA which is tagged with radioactive, molecule. It can search out complimentary DNA sequences from an organism.
Question 2.
Give a brief account of the tools of recombinant DNA technology.
Answer:
Key tools are :
1) Restriction enzymes :
Two enzymes responsible for restricting the growth of Bacteriophage in Escherichia coli were isolated in the year 1963. One of these added methyl groups to DNA and the other cut DNA. The latter was called restriction endonuclease. The first restriction endonuclease – Hind II which cut.
DNA molecules at a particular point by recognising a specific sequence of six base pairs, called recognition sequence for Hind II. Today, more than 900 restriction enzymes were isolated from over 200 strains of Bacteria, each of which recognises a different recognition sequence.
E CORI is a restriction enzyme in which, the first letter comes from the genus (Escherichia), and the second two letters from the species of the Prokaryotic cell [coli], the letter ‘R’ is derived from the name of strain. Roman numbers indicate the order in which the enzymes were isolated from that strain of Bacteria. Restriction enzymes belong to a larger class of enzymes called nucleases. They are of two types.
a) ExOnucleases which remove nucleotides from the ends of the DNA.
b) Endonucleases which make cuts at specific location with in the DNA.
Most restriction enzymes cut the two strands of DNA double helix at different locations. Such a cleavage is know as staggered cut. E CoRI recognises 5′ GAATT 3′ sites on the DNA and cuts it between G and A results in the formation of sticky ends or cohesive end pieces. This stickyness of the ends facilitates the action of enzyme DNA ligase.
Cloning vectors :
The DNA used as a carrier for transferring a fragment of foreign DNA into a suitable host is called vector. Vectors used for multiplying the foreign DNA sequences are called cloning vectors. Commonly used cloning vectors are plasmids, bacteriophages, cosmids. Plasmids are extrachromosomal circular DNA molecules present in almost all bacterial species. They are inheritable and carry few genes are easy to isolate and reintroduce into the bacterium (Host).
Features required to facilitate cloning into a vector:
a) Origin of replication:
(ori) This is a sequence from where replication starts and any piece of DNA when linked to this sequence can be made to replicate within host cells. It is also responsible for controlling the copy number of the linked DNA.
b) Selectable marker :
In addition to ‘ori’, the vector requires a selectable marker, which helps in identifying and eliminating non-transformants and selectively permitting the growth of the any transformants normally, the genes encoding resistance to antibiotics such as ampicillin, chloramphenicol, tetracycline or kanamycin etc., are useful selectable markers for E.Coli.
c) Cloning sites :
In order to link the alien DNA, the vector needs to have very few, preferably single recognition sites for the restriction enzymes.
d) Molecular weight:
The cloning vector should have low molecular weight.
e) Vectors for cloning genes in plants and animals:
The tumour inducing (Ti) plasmid of Agrobacterium tumifaciens has now been modified into a cloning vector such that it is no more pathogenic to plants. Similarly retroviruses have also been disarmed and are now used to deliver desirable genes into animal cells.
Intext Questions
Question 1.
Do Eukaryotic cells have restriction Endonucleases? Justify your answer.
Answer:
No. only prokaryotic cells have restriction Endonucleases. Today we know 900 or more restriction enzymes have been Isolated from over 230 strains of Bacteria.
![]()
Question 2.
Besides better aeration and mixing properties, what other advantages do stirred tank bioreactors have over shake flasks?
Answer:
- Small volumes of the culture can be withdrawn periodically.
- Air can be bubbled through the reactor.
- It has a control system that regulates the temperature and pH.
Question 3.
Can you recall Meiosis and indicate at what stage a recombinant DNA is made?
Answer:
Pachytene of Meiosis – I.
4. Describe briefly the following :
a) Origin of replication b) Bioreactors c) Down stream processing
Answer:
a) Origin of replication :
It is the specific DNA sequence which is responsible for initiating replication.
b) Bioreactors:
They are large vessels which are used for biological conversion of raw material into specific products.
c) Down stream processing :
Separation and purification of products is called down stream processing.
Question 5.
Explain briefly a) PCR b) Restriction enzymes and DNA c) Chitinase
Answer:
a) PCR:
Polymerase chain reaction: Is a biochemical technology in molecular biology to amplify a single or few copies of a piece of DNA generating thousands of copies of a particular DNA sequence.
b) Restriction enzymes and DNA :
Restriction enzymes are DNA cutting enzymes found in Bacteria because they cut with in the molecule.
c) Chitinase :
It is an enzyme obtained from the fungus, used in the digesting the bacterial cell walls.
Question 6.
Discuss with your teacher and find out how to distinguish between
a) Plasmid DNA and Chromosomal DNA
b) RNA and DNA
c) ExonuCleases and Endonucleases
a)
| Plasmid DNA | Chromosomal DNA |
| 1. Plasmid DNA is a small circular molecule found in Bacteria. | 1. It is main genetic material which is a circular, double stranded DNA |
| 2. It carry a few genes. | 2. It carry several genes. |
b)
| RNA | DNA |
| 1. It is made up of only polynucleotide chain. | 1. It is made up of 2 polynucleotide chains. |
| 2. Purines or pyTamidines do not exist in 1 : 1 ratio. | 2. Purines and pyramidines exist in 1 : 1 ratio. |
| 3. Nitrogen bases are Adenine, Guanine, uracil and cytosine. | 3. Nitrogen bases are Adenine, Guanine, Thymine and cytosine. |
c)
| Exonuclease | Endonuclease |
| 1. They remove nucleotides grom the ends of the DNA. | 1. They make cuts at specific positions with in the DNA. |
Question 7.
What does ‘H’ in’d’ and HI refer to in the enzyme Hind III?
Answer:
‘H’ – The genes from which it was isolated – Haemophilus
in – Influenza (the species name) .
d – Refers the bacterial strain from which enzyme was isolated.
III – Third isolated enzyme from the strain.
Question 8.
Restriction enzymes should not have more than one site of action in the cloning site of a vector. Comment.
Answer:
The presence of more than one recognition site for the same enzyme with in the vector will generate several fragments which will complicate the gene cloning. So vector needs to have very few, preferably single, recognition site for the commonly used restriction enzymes.
Question 9.
What does ‘competent’ refer to ‘incompetent cells’ used in transformation experiments?
Answer:
DNA is a hydrophilic molecule. It cannot pass through cell membranes. In order to force Bacteria to take up the plasmid, the bacterial cells must be made competent to take up DNA by treating them with a bivalent cation such as calcium which increaes the efficiency with which DNA enters the bacterium.
Question 10.
What is the significance of adding proteases at the time of Isolation of Genetic material (DNA)?
Answer:
Proteins can be removed by treatment with proteases.
Question 11.
While doing a PCR, ‘denaturation’ step is missed. What will be its effect on the process?
Answer:
The segment of DNA cannot be amplified.
Question 12.
What modification is done on the Ti plasmid of Agrobacterium tumefascians to convert it into a cloning vector?
Answer:
It is no more pathogenic to plants but is still able to use the mechanisms to deliver genes of our interest into a variety of plants.
Question 13.
What is meant by gene cloning?
Answer:
It is the replication of DNA fragments by the use of a self-replicating genetic material. It duplicates only individual genes of organism’s DNA.
![]()
Question 14.
Decide the ratio between ester bonds and hydrogen bonds that are broken in each palindromic sequence of DNA when treated with E coRI during the formation of sticky ends.
Answer:
1 : 4.